If you’re imagining an enhanced chakra balancing experience or more efficient digestion of your vitamin supplements, you will be a little disappointed in this latest news from the Imperial College of London (ICL). On the other hand, if you have damaged tissue, this discovery could make your recovery much easier. From a January 7, 2019 news item on phys.org,
Materials are widely used to help heal wounds: Collagen sponges help treat burns and pressure sores, and scaffold-like implants are used to repair bones. However, the process of tissue repair changes over time, so scientists are developing biomaterials that interact with tissues as healing takes place
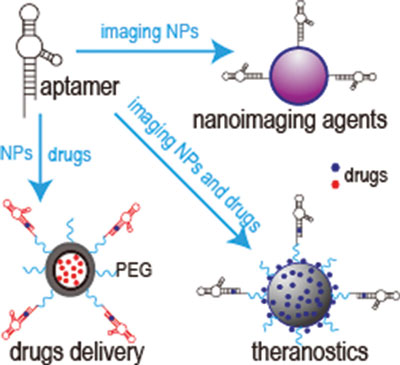
Now, Dr. Ben Almquist and his team at Imperial College London have created a new molecule that could change the way traditional materials work with the body. Known as traction force-activated payloads (TrAPs), their method lets materials talk to the body’s natural repair systems to drive healing.
The researchers say incorporating TrAPs into existing medical materials could revolutionise the way injuries are treated. Dr. Almquist, from Imperial’s Department of Bioengineering, said: “Our technology could help launch a new generation of materials that actively work with tissues to drive healing.”
A January 7, 2019 ICL press release (also on EurekAlert) by Caroline Brogan, which originated the news item, expands on the theme,
After an injury, cells ‘crawl’ through the collagen ‘scaffolds’ found in wounds, like spiders navigating webs. As they move, they pull on the scaffold, which activates hidden healing proteins that begin to repair injured tissue.
The researchers in the study designed TrAPs as a way to recreate this natural healing method. They folded the DNA segments into three-dimensional shapes known as aptamers that cling tightly to proteins. Then, they attached a customisable ‘handle’ that cells can grab onto on one end, before attaching the opposite end to a scaffold such as collagen.
During laboratory testing of their technique, they found that cells pulled on the TrAPs as they crawled through the collagen scaffolds. The pulling made the TrAPs unravel like shoelaces to reveal and activate the healing proteins. These proteins instruct the healing cells to grow and multiply
The researchers also found that by changing the cellular ‘handle’, they can change which type of cell can grab hold and pull, letting them tailor TrAPs to release specific therapeutic proteins based on which cells are present at a given point in time. In doing so, the TrAPs produce materials that can smartly interact with the correct type of cell at the correct time during wound repair.
This is the first time scientists have activated healing proteins using differing cell types in man-made materials. The technique mimics healing methods found in nature. Dr Almquist said: “Creatures from sea sponges to humans use cell movement to activate healing. Our approach mimics this by using the different cell varieties in wounds to drive healing.””
From lab to humans
This approach is adaptable to different cell types, so could be used in a variety of injuries such as fractured bones, scar tissue after heart attacks, and damaged nerves. New techniques are also desperately needed for patients whose wounds won’t heal despite current interventions, like diabetic foot ulcers, which are the leading cause of non-traumatic lower leg amputations.
TrAPs are relatively straightforward to create and are fully man-made, meaning they are easily recreated in different labs and can be scaled up to industrial quantities. Their adaptability also means they could help scientists create new methods for laboratory studies of diseases, stem cells, and tissue development.
Aptamers are currently used as drugs, meaning they are already proven safe and optimised for clinical use. Because TrAPs take advantage of aptamers that are safe for humans, they may be able to take a shorter path to the clinic than methods that start from ground zero.
Dr Almquist said: “TrAPs provide a flexible method of actively communicating with wounds, as well as key instructions when and where they are needed. This intelligent healing is useful during every phase of the healing process, has the potential to increase the body’s chance to recover, and has far-reaching uses on many different types of wounds. This technology could serve as a conductor of wound repair, orchestrating different cells over time to work together to heal damaged tissues.”
The researchers have made available an image and a video abstract illustrating their work,
By the way, the video was produced by www.animateyour.science (based in Adelaide, Australia) and they have a very interesting About page,
Our story
Your research is brilliant and novel. I’m sure of it. You might even be a pioneer in your field. But ask yourself honestly, is it enough? Is it truly enough to make a difference in the world?
My name is Tullio Rossi, and I founded Animate Your Science on my quest to make a positive impact on society through science.
During my Ph.D., I found that my peer-reviewed paper alone wasn’t cutting it. If I wanted to reach my peers, let alone the general public, I needed to communicate my findings in a fun and imaginative way.
This realization changed everything and inspired me to create “Lost at Sea,” an award-winning video that reached the hearts and minds of thousands of people.
The success of this first video blew my mind. And I got to thinking, maybe other scientists are lost at sea, so to speak. Maybe others want to reach the masses with their research, but just don’t know where to start.
This was the day Animate Your Science was born.
Why we do it
What we really want to do is bring science into society. That’s the true value of this company and the reason we believe in it.
We love science but we believe that, if not communicated properly, science is of limited use to society.
As scientists, it’s our privilege and duty to unearth these revelations and package them in a way that appeals to our peers as well as the general public.…
Getting back to TrAPS, here’s a link to and a citation for the paper,
Biologically Inspired, Cell‐Selective Release of Aptamer‐Trapped Growth Factors by Traction Forces by Anna Stejskalová, Nuria Oliva, Frances J. England, Benjamin D. Almquist. Advanced Materials DOI: https://doi.org/10.1002/adma.201806380 First published: 07 January 2019
This paper is open access.