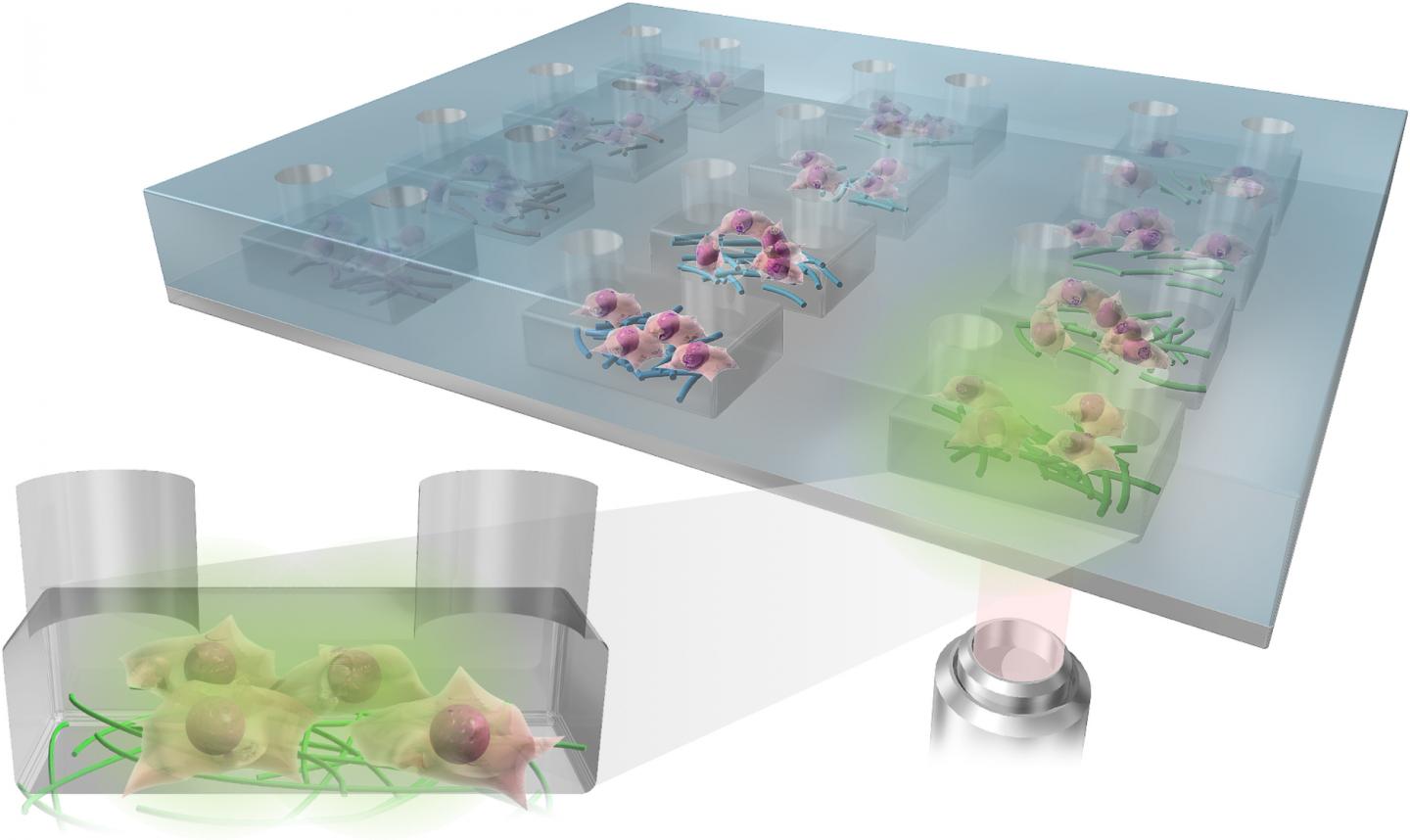
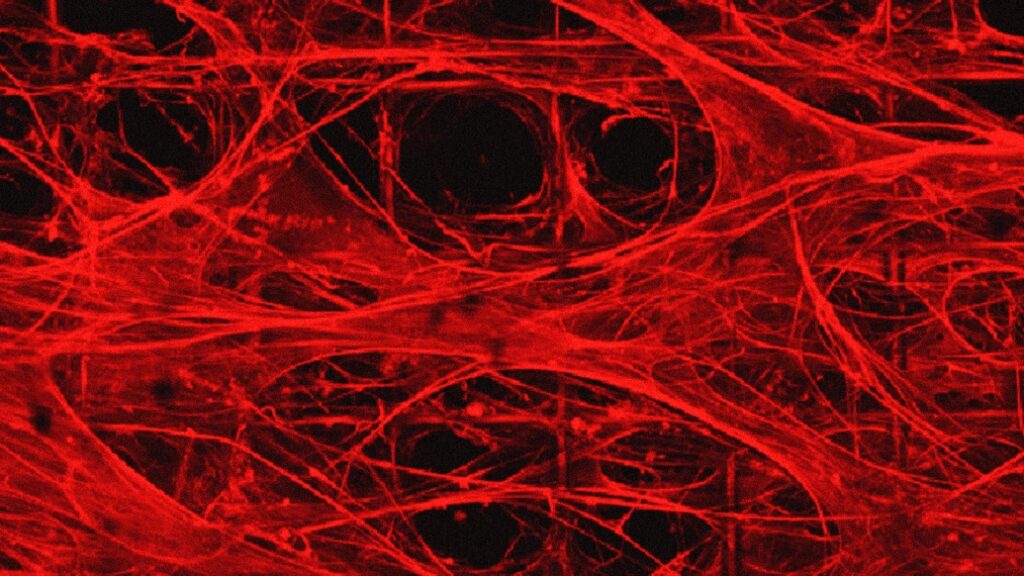
There’s a good (brief) description of how these fibres become organoids in the photo caption,
A July 13 ,2023 University of Michigan (Michigan Medicine) news release by Noah Fromson (also on EurekAlert) announces ‘kinder, gentler’ brain organoids. Coincidentally, these organoids more closely resemble human brains, Note: Links have been removed,
Researchers at University of Michigan developed a method to produce artificially grown miniature brains — called human brain organoids — free of animal cells that could greatly improve the way neurodegenerative conditions are studied and, eventually, treated.
Over the last decade of researching neurologic diseases, scientists have explored the use of human brain organoids as an alternative to mouse models. These self-assembled, 3D tissues derived from embryonic or pluripotent stem cells more closely model the complex brain structure compared to conventional two-dimensional cultures.
Until now, the engineered network of proteins and molecules that give structure to the cells in brain organoids, known as extracellular matrices, often used a substance derived from mouse sarcomas called Matrigel. That method suffers significant disadvantages, with a relatively undefined composition and batch-to-batch variability.
The latest U-M research, published in Annals of Clinical and Translational Neurology, offers a solution to overcome Matrigel’s weaknesses. Investigators created a novel culture method that uses an engineered extracellular matrix for human brain organoids — without the presence of animal components – and enhanced the neurogenesis of brain organoids compared to previous studies.
“This advancement in the development of human brain organoids free of animal components will allow for significant strides in the understanding of neurodevelopmental biology,” said senior author Joerg Lahann, Ph.D., director of the U-M Biointerfaces Institute and Wolfgang Pauli Collegiate Professor of Chemical Engineering at U-M.
“Scientists have long struggled to translate animal research into the clinical world, and this novel method will make it easier for translational research to make its way from the lab to the clinic.”
The foundational extracellular matrices of the research team’s brain organoids were comprised of human fibronectin, a protein that serves as a native structure for stem cells to adhere, differentiate and mature. They were supported by a highly porous polymer scaffold.
The organoids were cultured for months, while lab staff was unable to enter the building due to the COVID 19-pandemic.
Using proteomics, researchers found their brain organoids developed cerebral spinal fluid, a clear liquid that flows around healthy brain and spinal cords. This fluid more closely matched human adult CSF compared to a landmark study of human brain organoids developed in Matrigel.
“When our brains are naturally developing in utero, they are of course not growing on a bed of extracellular matrix produced by mouse cancer cells,” said first author Ayşe Muñiz, Ph.D., who was a graduate student in the U-M Macromolecular Science and Engineering Program at the time of the work.
“By putting cells in an engineered niche that more closely resembles their natural environment, we predicted we would observe differences in organoid development that more faithfully mimics what we see in nature.”
The success of these xenogeneic-free human brain organoids opens the door for reprogramming with cells from patients with neurodegenerative diseases, says co-author Eva Feldman, M.D., Ph.D., director of the ALS Center of Excellence at U-M and James W. Albers Distinguished Professor of Neurology at U-M Medical School.
“There is a possibility to take the stem cells from a patient with a condition such as ALS or Alzheimer’s and, essentially, build an avatar mini brain of that patients to investigate possible treatments or model how their disease will progress,” Feldman said. “These models would create another avenue to predict disease and study treatment on a personalized level for conditions that often vary greatly from person to person.”
Here’s a link to and a citation for the paper,
Engineered extracellular matrices facilitate brain organoids from human pluripotent stem cells by Ayşe J. Muñiz, Tuğba Topal, Michael D. Brooks, Angela Sze, Do Hoon Kim, Jacob Jordahl, Joe Nguyen, Paul H. Krebsbach, Masha G. Savelieff, Eva L. Feldman, Joerg Lahann. Annals of Clinical and Translational Neurology DOI: https://doi.org/10.1002/acn3.51820 First published: 07 June 2023
This paper is open access.