The academic paper for this latest research from Delft University of Technology (TU Delft, Netherlands), uses the term ‘bacterial sculptures,’ an intriguing idea that seems to have influenced the artistic illustration accompanying the research announcement.
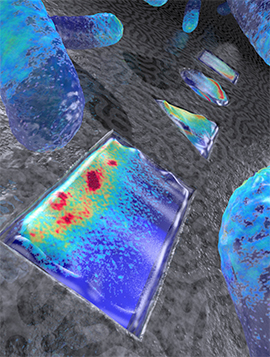
Artistic rendering live E.coli bacteria that have been shaped into a rectangle, triangle, circle, and square (from front to back). Colors indicate the density of the Min proteins that represent a snapshot in time (based on actual data), as these proteins oscillate back and forth within the bacterium, to determine the mid plane of the cell for cellular division.
Image credit: ‘Image Cees Dekker lab TU Delft / Tremani’
A June 22, 2015 news item on Nanowerk provides more insight into the research (Note: A link has been removed),
The E.coli bacterium, a very common resident of people’s intestines, is shaped as a tiny rod about 3 micrometers long. For the first time, scientists from the Kavli Institute of Nanoscience at Delft University have found a way to use nanotechnology to grow living E.coli bacteria into very different shapes: squares, triangles, circles, and even as letters spelling out ‘TU Delft’. They also managed to grow supersized E.coli with a volume thirty times larger than normal. These living oddly-shaped bacteria allow studies of the internal distribution of proteins and DNA in entirely new ways.
In this week’s Nature Nanotechnology (“Symmetry and scale orient Min protein patterns in shaped bacterial sculptures”), the scientists describe how these custom-designed bacteria still manage to perfectly locate ‘the middle of themselves’ for their cell division. They are found to do so using proteins that sense the cell shape, based on a mathematical principle proposed by computer pioneer Alan Turing in 1953.
A June 22, 2015 TU Delft press release, which originated the news item, expands on the theme,
Cell division
“If cells can’t divide properly, biological life wouldn’t be possible. Cells need to distribute their cell volume and genetic materials equally into their daughter cells to proliferate.”, says prof. Cees Dekker, “It is fascinating that even a unicellular organism knows how to divide very precisely. The distribution of certain proteins in the cell is key to regulating this, but how exactly do those proteins get that done?”
Turing
As the work of the Delft scientist exemplifies, the key here is a process discovered by the famous Alan Turing in 1953. Although Turing is mostly known for his role in deciphering the Enigma coding machine and the Turing Test, the impact of his ‘reaction-diffusion theory’ on biology might be even more spectacular. He predicted how patterns in space and time emerge as the result of only two molecular interactions – explaining for instance how a zebra gets its stripes, or how an embryo hand develops five fingers.
MinD and MinE
Such a Turing process also acts with proteins within a single cell, to regulate cell division. An E.coli cell uses two types of proteins, known as MinD and MinE, that bind and unbind again and again at the inner surface of the bacterium, thus oscillating back and forth from pole to pole within the bacterium every minute. “This results in a low average concentration of the protein in the middle and high concentrations at the ends, which drives the division machinery to the cell center”, says PhD-student Fabai Wu, who ran the experiments. “As our experiments show, the Turing patterns allow the bacterium to determine its symmetry axes and its center. This applies to many bacterial cell shapes that we custom-designed, such as squares, triangles and rectangles of many sizes. For fun, we even made ‘TUDelft’ and ‘TURING’ letters. Using computer simulations, we uncovered that the shape-sensing abilities are caused by simple Turing-type interactions between the proteins.”
Actual data for live E.coli bacteria that have been shaped into the letters TUDELFT.
The red color shows the cytosol contents of the cell, while the green color shows the density of the Min proteins, representing a snapshot in time, as these proteins oscillate back and forth within the bacterium to determine the mid plane of the cell for cellular division. The letters are about 5 micron high.
Image credit: ‘Fabai Wu, Cees Dekker lab at TU Delft’Spatial control for building synthetic cells
“Discovering this process is not only vital for our understanding of bacterial cell division – which is important in developing new strategies for antibiotics. But the approach will likely also be fruitful to figuring out how cells distribute other vital systems within a cell, such as chromosomes”, says Cees Dekker. “The ultimate goal in our research is to be able to completely build a living cell from artificial components, as that is the only way to really understand how life works. Understanding cell division – both the process that actually pinches off the cell into two daughters and the part that spatially regulates that machinery – is a major part of that.”
Here’s a link to and a citation for the paper,
Symmetry and scale orient Min protein patterns in shaped bacterial sculptures by Fabai Wu, Bas G. C. van Schie, Juan E. Keymer, & Cees Dekker. Nature Nanotechnology (2015) doi:10.1038/nnano.2015.126 Published online 22 June 2015
This paper is behind a paywall but there does seem to be another link (in the excerpt below) which gives you a free preview via ReadCube Access (according to the TU Delft press release),
The DOI for this paper will be 10.1038/nnano.2015.126. Once the paper is published electronically, the DOI can be used to retrieve the abstract and full text by adding it to the following url: http://dx.doi.org/
Enjoy!
