An August 15, 2023 news item on ScienceDaily breaks news from the University of Montréal,
Two molecular languages at the origin of life have been successfully recreated and mathematically validated, thanks to pioneering work by Canadian scientists at Université de Montréal.
…
Fascinating, non? An August 15, 2023 Université de Montréal news release (also on EurekAlert), which originated the news item, explaining how this leads to nanotechnology-enabled applications, Note: A link has been removed,
Published this week in the Journal of American Chemical Society, the breakthrough opens new doors for the development of nanotechnologies with applications ranging from biosensing, drug delivery and molecular imaging.
Living organisms are made up of billions of nanomachines and nanostructures that communicate to create higher-order entities able to do many essential things, such as moving, thinking, surviving and reproducing.
“The key to life’s emergence relies on the development of molecular languages – also called signalling mechanisms – which ensure that all molecules in living organisms are working together to achieve specific tasks,” said the study’s principal investigator, UdeM bioengineering professor Alexis Vallée-Bélisle.
In yeasts, for example, upon detecting and binding a mating pheromone, billions of molecules will communicate and coordinate their activities to initiate union, said Vallée-Bélisle, holder of a Canada Research Chair in Bioengineering and Bionanotechnology.
“As we enter the era of nanotechnology, many scientists believe that the key to designing and programming more complex and useful artificial nanosystems relies on our ability to understand and better employ molecular languages developed by living organisms,” he said.
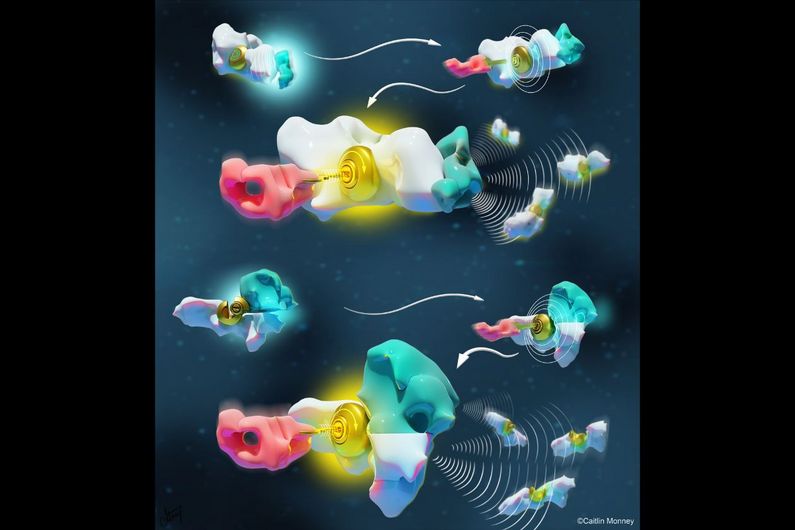
Two types of languages
One well-known molecular language is allostery. The mechanism of this language is “lock-and-key”: a molecule binds and modifies the structure of another molecule, directing it to trigger or inhibit an activity.
Another, lesser-known molecular language is multivalency, also known as the chelate effect. It works like a puzzle: as one molecule binds to another, it facilitates (or not) the binding of a third molecule by simply increasing its binding interface.
Although these two languages are observed in all molecular systems of all living organisms, it is only recently that scientists have started to understand their rules and principles – and so use these languages to design and program novel artificial nanotechnologies.
“Given the complexity of natural nanosystems, before now nobody was able to compare the basic rules, advantage or limitations of these two languages on the same system,” said Vallée-Bélisle.
To do so, his doctoral student Dominic Lauzon, first author of the study, had the idea of creating a DNA-based molecular system that could function using both languages. “DNA is like Lego bricks for nanoengineers,” said Lauzon. “It’s a remarkable molecule that offers simple, programmable and easy-to-use chemistry.”
Simple mathematical equations to detect antibodies
The researchers found that simple mathematical equations could well describe both languages, which unravelled the parameters and design rules to program the communication between molecules within a nanosystem.
For example, while the multivalent language enabled control of both the sensitivity and cooperativity of the activation or deactivation of the molecules, the corresponding allosteric translation only enabled control of the sensitivity of the response.
With this new understanding at hand, the researchers used the language of multivalency to design and engineer a programmable antibody sensor that allows the detection of antibodies over different ranges of concentration.
“As shown with the recent pandemic, our ability to precisely monitor the concentration of antibodies in the general population is a powerful tool to determine the people’s individual and collective immunity,” said Vallée-Bélisle.
In addition to expanding the synthetic toolbox to create the next generation of nanotechnology, the scientist’s discovery also shines a light on why some natural nanosystems may have selected one language over another to communicate chemical information.
Here’s a link to and a citation for the paper,
Programing Chemical Communication: Allostery vs Multivalent Mechanism by Dominic Lauzon and Alexis Vallée-Bélisle. J. Am. Chem. Soc. 2023, XXXX, XXX, XXX-XXX DOI: https://doi.org/10.1021/jacs.3c04045 Online Publication Date: August 15, 2023 © 2023 American Chemical Society
This paper is behind a paywall.
![[downloaded from http://pubs.acs.org/doi/abs/10.1021/acs.langmuir.6b01743]](http://www.frogheart.ca/wp-content/uploads/2016/07/KazanUni_Oil-degradingBacteria.gif)
![Fig. 1: A Vigna radiata seeds. b Reddish brown solution of silver nanoparticles formed after 3 h due to reduction of silver ions [downloaded from http://link.springer.com/article/10.1007/s13204-015-0418-6]](http://www.frogheart.ca/wp-content/uploads/2016/07/India_SilverNanoparrticles_MungBean.gif)