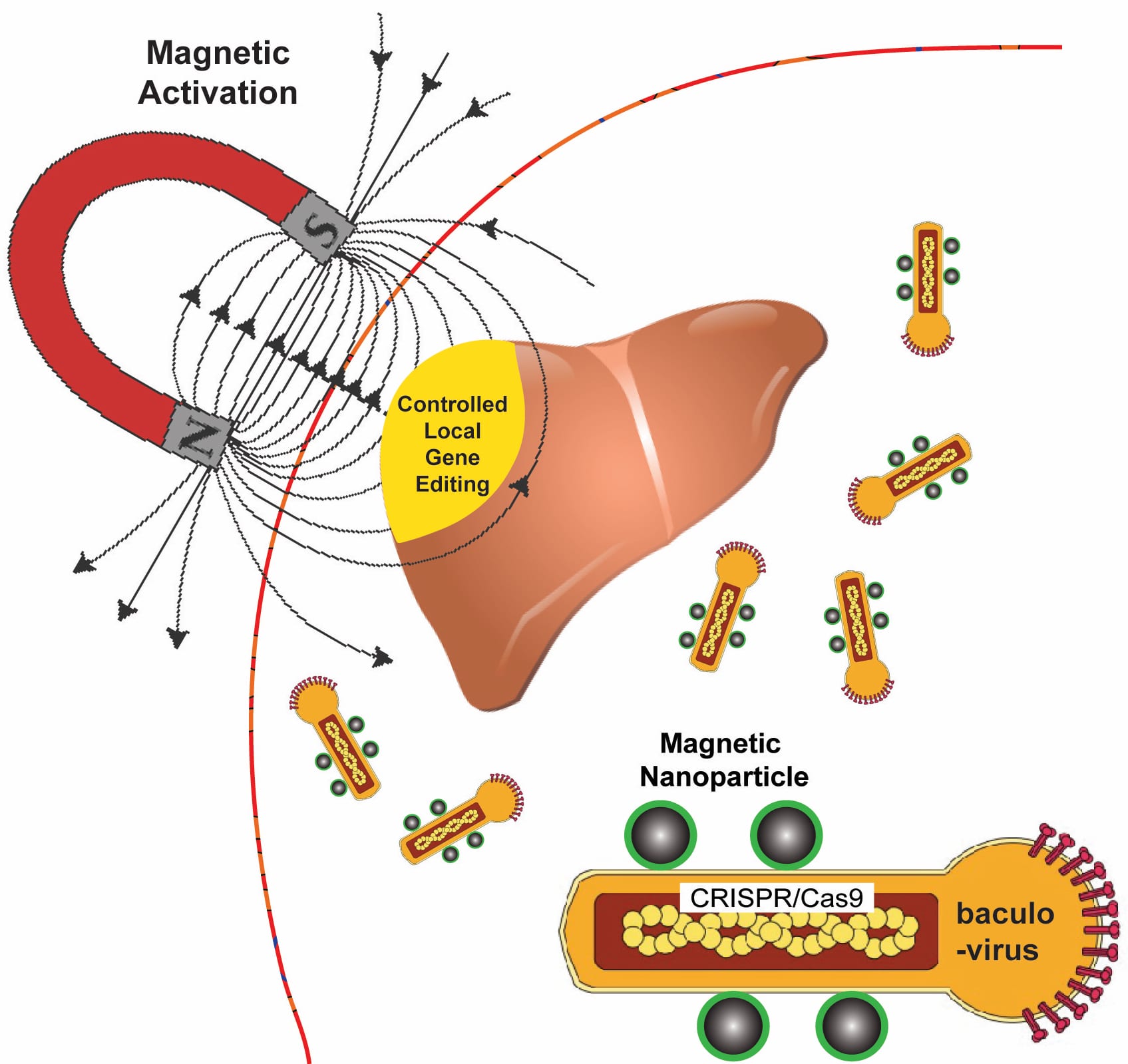
Rice University bioengineers use a magnetic field to activate nanoparticle-attached baculoviruses in a tissue. The viruses, which normally infect alfalfa looper moths, are modified to deliver gene-editing DNA code only to cells that are targeted with magnetic field-induced local transduction. Courtesy of the Laboratory of Biomolecular Engineering and Nanomedicine
Kudos to whomever put that diagram together! That’s a lot of well conveyed information.
Now for the details about how this technology might save lives. From a November 13, 2018 news item on Nanowerk,
A new technology that relies on a moth-infecting virus and nanomagnets could be used to edit defective genes that give rise to diseases like sickle cell, muscular dystrophy and cystic fibrosis.
Rice University bioengineer Gang Bao has combined magnetic nanoparticles with a viral container drawn from a particular species of moth to deliver CRISPR/Cas9 payloads that modify genes in a specific tissue or organ with spatial control.
A November 12, 2018 Rice University news release (also on EurekAlert published on November 13, 2018), which originated the news item, provides detail,
Because magnetic fields are simple to manipulate and, unlike light, pass easily through tissue, Bao and his colleagues want to use them to control the expression of viral payloads in target tissues by activating the virus that is otherwise inactivated in blood.
The research appears in Nature Biomedical Engineering. In nature, CRISPR/Cas9 bolsters microbes’ immune systems by recording the DNA of invaders. That gives microbes the ability to recognize and attack returning invaders, but scientists have been racing to adapt CRISPR/Cas9 to repair mutations that cause genetic diseases and to manipulate DNA in laboratory experiments.
CRISPR/Cas9 has the potential to halt hereditary disease – if scientists can get the genome-editing machinery to the right cells inside the body. But roadblocks remain, especially in delivering the gene-editing payloads with high efficiency.
Bao said it will be necessary to edit cells in the body to treat many diseases. “But efficiently delivering genome-editing machinery into target tissue in the body with spatial control remains a major challenge,” Bao said. “Even if you inject the viral vector locally, it can leak to other tissues and organs, and that could be dangerous.”
The delivery vehicle developed by Bao’s group is based on a virus that infects Autographa californica, aka the alfalfa looper, a moth native to North America. The cylindrical baculovirus vector (BV), the payload-carrying part of the virus, is considered large at up to 60 nanometers in diameter and 200-300 nanometers in length. That’s big enough to transport more than 38,000 base pairs of DNA, which is enough to supply multiple gene-editing units to a target cell, Bao said.
He said the inspiration to combine BV and magnetic nanoparticles came from discussions with Rice postdoctoral researcher and co-lead author Haibao Zhu, who learned about the virus during a postdoctoral stint in Singapore but knew nothing about magnetic nanoparticles until he joined the Bao lab. The Rice team had previous experience using iron oxide nanoparticles and an applied magnetic field to open blood vessel walls just enough to let large-molecule drugs pass through.
“We really didn’t know if this would work for gene editing or not, but we thought, ‘worth a shot,'” Bao said.
The researchers use the magnetic nanoparticles to activate BV and deliver gene-editing payloads only where they’re needed. To do this, they take advantage of an immune-system protein called C3 that normally inactivates baculoviruses.
“If we combine BV with magnetic nanoparticles, we can overcome this deactivation by applying the magnetic field,” Bao said. “The beauty is that when we deliver it, gene editing occurs only at the tissue, or the part of the tissue, where we apply the magnetic field.”
Application of the magnetic field allows BV transduction, the payload-delivery process that introduces gene-editing cargo into the target cell. The payload is also DNA, which encodes both a reporter gene and the CRISPR/Cas9 system.
In tests, the BV was loaded with green fluorescent proteins or firefly luciferase. Cells with the protein glowed brightly under a microscope, and experiments showed the magnets were highly effective at targeted delivery of BV cargoes in both cell cultures and lab animals.
Bao noted his and other labs are working on the delivery of CRISPR/Cas9 with adeno-associated viruses (AAV), but he said BV’s capacity for therapeutic cargo is roughly eight times larger. “However, it is necessary to make BV transduction into target cells more efficient,” he said.
Here’s a link to and a citation for the paper,
Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets by Haibao Zhu, Linlin Zhang, Sheng Tong, Ciaran M. Lee, Harshavardhan Deshmukh, & Gang Bao. Nature Biomedical Engineering (2018) DOI: https://doi.org/10.1038/s41551-018-0318-7 Published: 12 November 2018
This paper is behind a paywall.