This* story actually started in 2018 with an August 1, 2018 Harvard University news release (h/t Aug. 1, 2018 news item on phys.org) by Leslie Brownell announcing molecular and synthetic biology educational kits that been tested in the classroom. (In 2019, a new kit was released but more about that later.)
As biologists have probed deeper into the molecular and genetic underpinnings of life, K-12 schools have struggled to provide a curriculum that reflects those advances. Hands-on learning is known to be more engaging and effective for teaching science to students, but even the most basic molecular and synthetic biology experiments require equipment far beyond an average classroom’s budget, and often involve the use of bacteria and other substances that can be difficult to manage outside a controlled lab setting.
Now, a collaboration between the Wyss Institute at Harvard University, MIT [Massachusetts Institute of Technology], and Northwestern University has developed BioBits, new educational biology kits that use freeze-dried cell-free (FD-CF) reactions to enable students to perform a range of simple, hands-on biological experiments. The BioBits kits introduce molecular and synthetic biology concepts without the need for specialized lab equipment, at a fraction of the cost of current standard experimental designs. The kits are described in two papers published in Science Advances [2018].
“The main motivation in developing these kits was to give students fun activities that allow them to actually see, smell, and touch the outcomes of the biological reactions they’re doing at the molecular level,” said Ally Huang, a co-first author on both papers who is an MIT graduate student in the lab of Wyss Founding Core Faculty member Jim Collins, Ph.D. “My hope is that they will inspire more kids to consider a career in STEM [science, technology, engineering, and math] and, more generally, give all students a basic understanding of how biology works, because they may one day have to make personal or policy decisions based on modern science.”
Synthetic and molecular biology frequently make use of the cellular machinery found in E. coli bacteria to produce a desired protein. But this system requires that the bacteria be kept alive and contained for an extended period of time, and involves several complicated preparation and processing steps. The FD-CF reactions pioneered in Collins’ lab for molecular manufacturing, when combined with innovations from the lab of Michael Jewett, Ph.D. at Northwestern University, offer a solution to this problem by removing bacteria from the equation altogether.
“You can think of it like opening the hood of a car and taking the engine out: we’ve taken the ‘engine’ that drives protein production out of a bacterial cell and given it the fuel it needs, including ribosomes and amino acids, to create proteins from DNA outside of the bacteria itself,” explained Jewett, who is the Charles Deering McCormick Professor of Teaching Excellence at Northwestern University’s McCormick School of Engineering and co-director of Northwestern’s Center for Synthetic Biology, and co-corresponding author of both papers. This collection of molecular machinery is then freeze-dried into pellets so that it becomes shelf-stable at room temperature. To initiate the transcription of DNA into RNA and the translation of that RNA into a protein, a student just needs to add the desired DNA and water to the freeze-dried pellets.
The researchers designed a range of molecular experiments that can be performed using this system, and coupled each of them to a signal that the students can easily detect with their sense of sight, smell, or touch. The first, called BioBits Bright, contains six different freeze-dried DNA templates that each encode a protein that fluoresces a different color when illuminated with blue light. To produce the proteins, students simply add these DNA templates and water to the FD-CF machinery and put the reactions in an inexpensive incubator (~$30) for several hours, and then view them with a blue light illuminator (~$15). The students can also design their own experiments to produce a desired collection of colors that they can then arrange into a visual image, a bit like using a Light Brite ©. “Challenging the students to build their own in vitro synthetic programs also allows educators to start to talk about how synthetic biologists might control biology to make important products, such as medicines or chemicals,” explained Jessica Stark, an NSF Graduate Research Fellow in the Jewett lab at Northwestern University who is co-first author on both papers.
An expansion of the BioBits Bright kit, called BioBits Explorer, includes experiments that engage the senses of smell and touch and allow students to probe their environment using designer synthetic biosensors. In the first experiment, the FD-CF reaction pellets contain a gene that drives the conversion of isoamyl alcohol to isoamyl acetate, a compound that produces a strong banana odor. In the second experiment, the FD-CF reactions contain a gene coding for the enzyme sortase, which recognizes and links specific segments of proteins in a liquid solution together to form a squishy, semi-solid hydrogel, which the students can touch and manipulate. The third module uses another Wyss technology, the toehold switch sensor, to identify DNA extracted from a banana or a kiwi. The sensors are hairpin-shaped RNA molecules designed such that when they bind to a “trigger” RNA, they spring open and reveal a genetic sequence that produces a fluorescent protein. When fruit DNA is added to the sensor-containing FD-CF pellets, only the sensors that are designed to open in the presence of each fruit’s RNA will produce the fluorescent protein.
The researchers tested their BioBits kits in the Chicago Public School system, and demonstrated that students and teachers were able to perform the experiments in the kits with the same success as trained synthetic biology researchers. In addition to refining the kits’ design so that they can one day provide them to classrooms around the world, the authors hope to create an open-source online database where teachers and students can share their results and ideas for ways to modify the kits to explore different biological questions.
“Synthetic biology is going to be one of the defining technologies of the century, and yet it has been challenging to teach the fundamental concepts of the field in K-12 classrooms given that such efforts often require expensive, complicated equipment,” said Collins, who is a co-corresponding author of both papers and also the Termeer Professor of Medical Engineering & Science at MIT. “We show that it is possible to use freeze-dried, cell-free extracts along with freeze-dried synthetic biology components to conduct innovative educational experiments in classrooms and other low-resource settings. The BioBits kits enable us to expose young kids, older kids, and even adults to the wonders of synthetic biology and, as a result, are poised to transform science education and society.
“All scientists are passionate about what they do, and we are frustrated by the difficulty our educational system has had in inciting a similar level of passion in young people. This BioBits project demonstrates the kind of out-of-the-box thinking and refusal to accept the status quo that we value and cultivate at the Wyss Institute, and we all hope it will stimulate young people to be intrigued by science,” said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at Harvard Medical School (HMS) and the Vascular Biology Program at Boston Children’s Hospital, as well as Professor of Bioengineering at Harvard’s John A. Paulson School of Engineering and Applied Sciences (SEAS). “It’s exciting to see this project move forward and become available to biology classrooms worldwide and, hopefully some of these students will pursue a path in science because of their experience.”
Additional authors of the papers include Peter Nguyen, Ph.D., Nina Donghia, and Tom Ferrante from the Wyss Institute; Melissa Takahashi, Ph.D. and Aaron Dy from MIT; Karen Hsu and Rachel Dubner from Northwestern University; Keith Pardee, Ph.D., Assistant Professor at the University of Toronto; and a number of teachers and students in the Chicago school system including: Mary Anderson, Ada Kanapskyte, Quinn Mucha, Jessica Packett, Palak Patel, Richa Patel, Deema Qaq, Tyler Zondor, Julie Burke, Tom Martinez, Ashlee Miller-Berry, Aparna Puppala, Kara Reichert, Miriam Schmid, Lance Brand, Lander Hill, Jemima Chellaswamy, Nuhie Faheem, Suzanne Fetherling, Elissa Gong, Eddie Marie Gonzales, Teresa Granito, Jenna Koritsaris, Binh Nguyen, Sujud Ottman, Christina Palffy, Angela Patel, Sheila Skweres, Adriane Slaton, and TaRhonda Woods.
This research was supported by the Army Research Office, the National Science Foundation, the Air Force Research Laboratory Center of Excellence Grant, The Defense Threat Reduction Agency Grant, the David and Lucile Packard Foundation, the Camille Dreyfus Teacher-Scholar Program, the Wyss Institute at Harvard University, the Paul G. Allen Frontiers Group, The Air Force Office of Scientific Research, and the Natural Sciences and Engineering Council of Canada. [emphases mine]
Well, that list of funding agencies is quite interesting. The US Army and Air Force but not the Navy? As for what the Natural Sciences and Engineering Council of Canada is doing on that list, I can only imagine why.
This is what they were doing in 2018,
Now for the latest update, a May 7, 2019 news item on phys.org announces the BioBits Kits have been expanded,
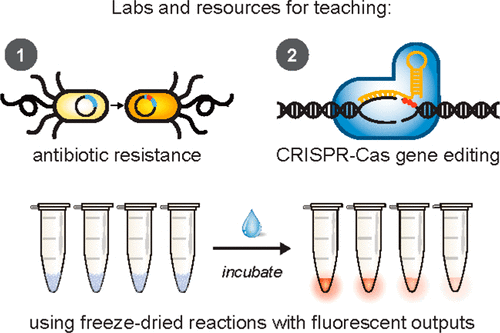
How can high school students learn about a technology as complex and abstract as CRISPR? It’s simple: just add water.
A Northwestern University-led team has developed BioBits, a suite of hands-on educational kits that enable students to perform a range of biological experiments by adding water and simple reagents to freeze-dried cell-free reactions. The kits link complex biological concepts to visual, fluorescent readouts, so students know—after a few hours and with a single glance—the results of their experiments.
A May 7, 2019 Northwestern University news release (also on EurekAlert and received via email) by Amanda Morris, which originated the news item, provides more details,
After launching BioBits last summer, the researchers are now expanding the kit to include modules for CRISPR [clustered regularly interspaced short palindromic repeats] and antibiotic resistance. A small group of Chicago-area teachers and high school students just completed the first pilot study for these new modules, which include interactive experiments and supplementary materials exploring ethics and strategies.
“After we unveiled the first kits, we next wanted to tackle current topics that are important for society,” said Northwestern’s Michael Jewett, principal investigator of the study. “That led us to two areas: antibiotic resistance and gene editing.”
Called BioBits Health, the new kits and pilot study are detailed in a paper published today (May 7 [2019]) in the journal ACS Synthetic Biology.
Jewett is a professor of chemical and biological engineering in Northwestern’s McCormick School of Engineering and co-director of Northwestern’s Center for Synthetic Biology. Jessica Stark, a graduate student in Jewett’s laboratory, led the study.
Test in a tube
Instead of using live cells, the BioBits team removed the essential cellular machinery from inside the cells and freeze-dried them for shelf stability. Keeping cells alive and contained for an extended period of time involves several complicated, time-consuming preparation and processing steps as well as expensive equipment. Freeze-dried cell-free reactions bypass those complications and costs.
“These are essentially test-tube biological reactions,” said Stark, a National Science Foundation graduate research fellow. “We break the cells open and use their guts, which still contain all of the necessary biological machinery to carry out a reaction. We no longer need living cells to demonstrate biology.”
This method to harness biological systems without intact, living cells became possible over the last two decades thanks to multiple innovations, including many in cell-free synthetic biology by Jewett’s lab. Not only are these experiments doable in the classroom, they also only cost pennies compared to standard high-tech experimental designs.
“I’m hopeful that students get excited about engineering biology and want to learn more,” Jewett said.
Conquering CRISPR
One of the biggest scientific breakthroughs of the past decade, CRISPR (pronounced “crisper”) stands for Clustered Regularly Interspaced Short Palindromic Repeats. The powerful gene-editing technology uses enzymes to cut DNA in precise locations to turn off or edit targeted genes. It could be used to halt genetic diseases, develop new medicines, make food more nutritious and much more.
BioBits Health uses three components required for CRISPR: an enzyme called the Cas9 protein, a target DNA sequence encoding a fluorescent protein and an RNA molecule that targets the fluorescent protein gene. When students add all three components — and water — to the freeze-dried cell-free system, it creates a reaction that edits, or cuts, the DNA for the fluorescent protein. If the DNA is cut, the system does not glow. If the DNA is not cut, the fluorescent protein is made, and the system glows fluorescent.
“We have linked this abstract, really advanced biological concept to the presence or absence of a fluorescent protein,” Stark said. “It’s something students can see, something they can visually understand.”
The curriculum also includes activities that challenge students to consider the ethical questions and dilemmas surrounding the use of gene-editing technologies.
“There is a lot of excitement about being able to edit genomes with these technologies,” Jewett said. “BioBits Health calls attention to a lot of important questions — not only about how CRISPR technology works but about ethics that society should be thinking about. We hope that this promotes a conversation and dialogue about such technologies.”
Reducing resistance
Jewett and Stark are both troubled by a prediction that, by the year 2050, drug-resistant bacterial infections could outpace cancer as a leading cause of death. This motivated them to help educate the future generation of scientists about how antibiotic resistance emerges and inspire them to take actions that could help limit the emergence of resistant bacteria.
In this module, students run two sets of reactions to produce a glowing fluorescent protein — one set with an antibiotic resistance gene and one set without. Students then add antibiotics. If the experiment glows, the fluorescent protein has been made, and the reaction has become resistant to antibiotics. If the experiment does not glow, then the antibiotic has worked.“Because we’re using cell-free systems rather than organisms, we can demonstrate drug resistance in a way that doesn’t create drug-resistant bacteria,” Stark explained. “We can demonstrate these concepts without the risks.”
A supporting curriculum piece challenges students to brainstorm and research strategies for slowing the rate of emerging antibiotic resistant strains.
Part of something cool
After BioBits was launched in summer 2018, 330 schools from around the globe requested prototype kits for their science labs. The research team, which includes members from Northwestern and MIT, has received encouraging feedback from teachers, students and parents.
“The students felt like scientists and doctors by touching and using the laboratory materials provided during the demo,” one teacher said. “Even the students who didn’t seem engaged were secretly paying attention and wanted to take their turn pipetting. They knew they were part of something really cool, so we were able to connect with them in a way that was new to them.”
“My favorite part was using the equipment,” a student said. “It was a fun activity that immerses you into what top scientists are currently doing.”
###
The study, “BioBits Health: Classroom activities exploring engineering, biology and human health with fluorescent readouts,” was supported by the Army Research Office (award number W911NF-16-1-0372), the National Science Foundation (grant numbers MCB-1413563 and MCB-1716766), the Air Force Research Laboratory Center of Excellence (grant number FA8650-15-2-5518), the Defense Threat Reduction Agency (grant number HDTRA1-15-10052/P00001), the Department of Energy (grant number DE-SC0018249), the Human Frontiers Science Program (grant number RGP0015/2017), the David and Lucile Packard Foundation, the Office of Energy Efficiency and Renewable Energy (grant number DE-EE008343) and the Camille Dreyfus Teacher-Scholar Program. [emphases mine]
This is an image you’ll find in the abstract for the 2019 paper,
Here are links and citations for the 2018 papers and the 2019 paper,
BioBits™ Explorer: A modular synthetic biology education kit by Ally Huang, Peter Q. Nguyen, Jessica C. Stark, Melissa K. Takahashi, Nina Donghia, Tom Ferrante, Aaron J. Dy, Karen J. Hsu, Rachel S. Dubner, Keith Pardee, Michael C. Jewett, and James J. Collins. Science Advances 01 Aug 2018: Vol. 4, no. 8, eaat5105 DOI: 10.1126/sciadv.aat5105
BioBits™ Bright: A fluorescent synthetic biology education kit by Jessica C. Stark, Ally Huang, Peter Q. Nguyen, Rachel S. Dubner, Karen J. Hsu, Thomas C. Ferrante, Mary Anderson, Ada Kanapskyte, Quinn Mucha, Jessica S. Packett, Palak Patel, Richa Patel, Deema Qaq, Tyler Zondor, Julie Burke, Thomas Martinez, Ashlee Miller-Berry, Aparna Puppala, Kara Reichert, Miriam Schmid, Lance Brand, Lander R. Hill, Jemima F. Chellaswamy, Nuhie Faheem, Suzanne Fetherling, Elissa Gong, Eddie Marie Gonzalzles, Teresa Granito, Jenna Koritsaris, Binh Nguyen, Sujud Ottman, Christina Palffy, Angela Patel, Sheila Skweres, Adriane Slaton, TaRhonda Woods, Nina Donghia, Keith Pardee, James J. Collins, and Michael C. Jewett. Science Advances 01 Aug 2018: Vol. 4, no. 8, eaat5107 DOI: 10.1126/sciadv.aat5107
BioBits Health: Classroom Activities Exploring Engineering, Biology, and Human Health with Fluorescent Readouts by Jessica C. Stark, Ally Huang, Karen J. Hsu, Rachel S. Dubner, Jason Forbrook, Suzanne Marshalla, Faith Rodriguez, Mechelle Washington, Grant A. Rybnicky, Peter Q. Nguyen, Brenna Hasselbacher, Ramah Jabri, Rijha Kamran, Veronica Koralewski, Will Wightkin, Thomas Martinez, and Michael C. Jewett. ACS Synth. Biol., Article ASAP
DOI: 10.1021/acssynbio.8b00381 Publication Date (Web): March 29, 2019
Copyright © 2019 American Chemical Society
Both of the 2018 papers appear to be open access while the 2019 paper is behind a paywall.
Should you be interested in acquiring a BioBits kit, you can check out the BioBits website. As for ‘conguering’ CRISPR, do we really need to look at it that way? Maybe a more humble appraoch could work just as well or even better, eh?
*’is’ removed from sentence on May 9, 2019.