Before getting to the announcement, here’s a little background from Dexter Johnson’s Feb. 21, 2017 posting on his NanoClast blog (on the IEEE [Institute of Electrical and Electronics Engineers] website; Note: Links have been removed),
Ever since the 1980s, when Gerd Binnig of IBM first heard that “beautiful noise” made by the tip of the first scanning tunneling microscope (STM) dragging across the surface of an atom, and he later developed the atomic force microscope (AFM), these microscopy tools have been the bedrock of nanotechnology research and development.
AFMs have continued to evolve over the years, and at one time, IBM even looked into using them as the basis of a memory technology in the company’s Millipede project. Despite all this development, AFMs have remained bulky and expensive devices, costing as much as $50,000 [or more].
…
Now, here’s the announcement in a Feb. 15, 2017 news item on Nanowerk,
Researchers at The University of Texas at Dallas have created an atomic force microscope on a chip, dramatically shrinking the size — and, hopefully, the price tag — of a high-tech device commonly used to characterize material properties.
“A standard atomic force microscope is a large, bulky instrument, with multiple control loops, electronics and amplifiers,” said Dr. Reza Moheimani, professor of mechanical engineering at UT Dallas. “We have managed to miniaturize all of the electromechanical components down onto a single small chip.”
A Feb. 15, 2017 University of Texas at Dallas news release, which originated the news item, provides more detail,
An atomic force microscope (AFM) is a scientific tool that is used to create detailed three-dimensional images of the surfaces of materials, down to the nanometer scale — that’s roughly on the scale of individual molecules.
The basic AFM design consists of a tiny cantilever, or arm, that has a sharp tip attached to one end. As the apparatus scans back and forth across the surface of a sample, or the sample moves under it, the interactive forces between the sample and the tip cause the cantilever to move up and down as the tip follows the contours of the surface. Those movements are then translated into an image.
“An AFM is a microscope that ‘sees’ a surface kind of the way a visually impaired person might, by touching. You can get a resolution that is well beyond what an optical microscope can achieve,” said Moheimani, who holds the James Von Ehr Distinguished Chair in Science and Technology in the Erik Jonsson School of Engineering and Computer Science. “It can capture features that are very, very small.”
The UT Dallas team created its prototype on-chip AFM using a microelectromechanical systems (MEMS) approach.
“A classic example of MEMS technology are the accelerometers and gyroscopes found in smartphones,” said Dr. Anthony Fowler, a research scientist in Moheimani’s Laboratory for Dynamics and Control of Nanosystems and one of the article’s co-authors. “These used to be big, expensive, mechanical devices, but using MEMS technology, accelerometers have shrunk down onto a single chip, which can be manufactured for just a few dollars apiece.”
The MEMS-based AFM is about 1 square centimeter in size, or a little smaller than a dime. It is attached to a small printed circuit board, about half the size of a credit card, which contains circuitry, sensors and other miniaturized components that control the movement and other aspects of the device.
Conventional AFMs operate in various modes. Some map out a sample’s features by maintaining a constant force as the probe tip drags across the surface, while others do so by maintaining a constant distance between the two.
“The problem with using a constant height approach is that the tip is applying varying forces on a sample all the time, which can damage a sample that is very soft,” Fowler said. “Or, if you are scanning a very hard surface, you could wear down the tip,”
The MEMS-based AFM operates in “tapping mode,” which means the cantilever and tip oscillate up and down perpendicular to the sample, and the tip alternately contacts then lifts off from the surface. As the probe moves back and forth across a sample material, a feedback loop maintains the height of that oscillation, ultimately creating an image.
“In tapping mode, as the oscillating cantilever moves across the surface topography, the amplitude of the oscillation wants to change as it interacts with sample,” said Dr. Mohammad Maroufi, a research associate in mechanical engineering and co-author of the paper. “This device creates an image by maintaining the amplitude of oscillation.”
Because conventional AFMs require lasers and other large components to operate, their use can be limited. They’re also expensive.
“An educational version can cost about $30,000 or $40,000, and a laboratory-level AFM can run $500,000 or more,” Moheimani said. “Our MEMS approach to AFM design has the potential to significantly reduce the complexity and cost of the instrument.
“One of the attractive aspects about MEMS is that you can mass produce them, building hundreds or thousands of them in one shot, so the price of each chip would only be a few dollars. As a result, you might be able to offer the whole miniature AFM system for a few thousand dollars.”
A reduced size and price tag also could expand the AFMs’ utility beyond current scientific applications.
“For example, the semiconductor industry might benefit from these small devices, in particular companies that manufacture the silicon wafers from which computer chips are made,” Moheimani said. “With our technology, you might have an array of AFMs to characterize the wafer’s surface to find micro-faults before the product is shipped out.”
The lab prototype is a first-generation device, Moheimani said, and the group is already working on ways to improve and streamline the fabrication of the device.
“This is one of those technologies where, as they say, ‘If you build it, they will come.’ We anticipate finding many applications as the technology matures,” Moheimani said.
In addition to the UT Dallas researchers, Michael Ruppert, a visiting graduate student from the University of Newcastle in Australia, was a co-author of the journal article. Moheimani was Ruppert’s doctoral advisor.
So, an AFM that could cost as much as $500,000 for a laboratory has been shrunk to this size and become far less expensive,
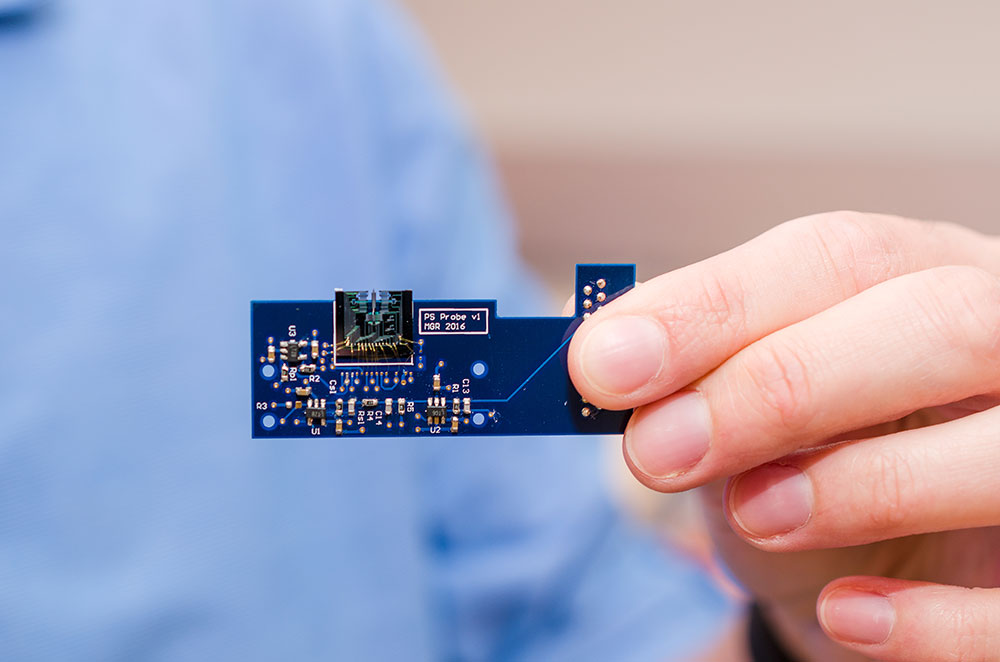
A MEMS-based atomic force microscope developed by engineers at UT Dallas is about 1 square centimeter in size (top center). Here it is attached to a small printed circuit board that contains circuitry, sensors and other miniaturized components that control the movement and other aspects of the device. Courtesy: University of Texas at Dallas
Of course, there’s still more work to be done as you’ll note when reading Dexter’s Feb. 21, 2017 posting where he features answers to questions he directed to the researchers.
Here’s a link to and a citation for the paper,
On-Chip Dynamic Mode Atomic Force Microscopy: A Silicon-on-Insulator MEMS Approach by Michael G. Ruppert, Anthony G. Fowler, Mohammad Maroufi, S. O. Reza Moheimani. IEEE Journal of Microelectromechanical Systems Volume: 26 Issue: 1 Feb. 2017 DOI: 10.1109/JMEMS.2016.2628890 Date of Publication: 06 December 2016
This paper is behind a paywall.


