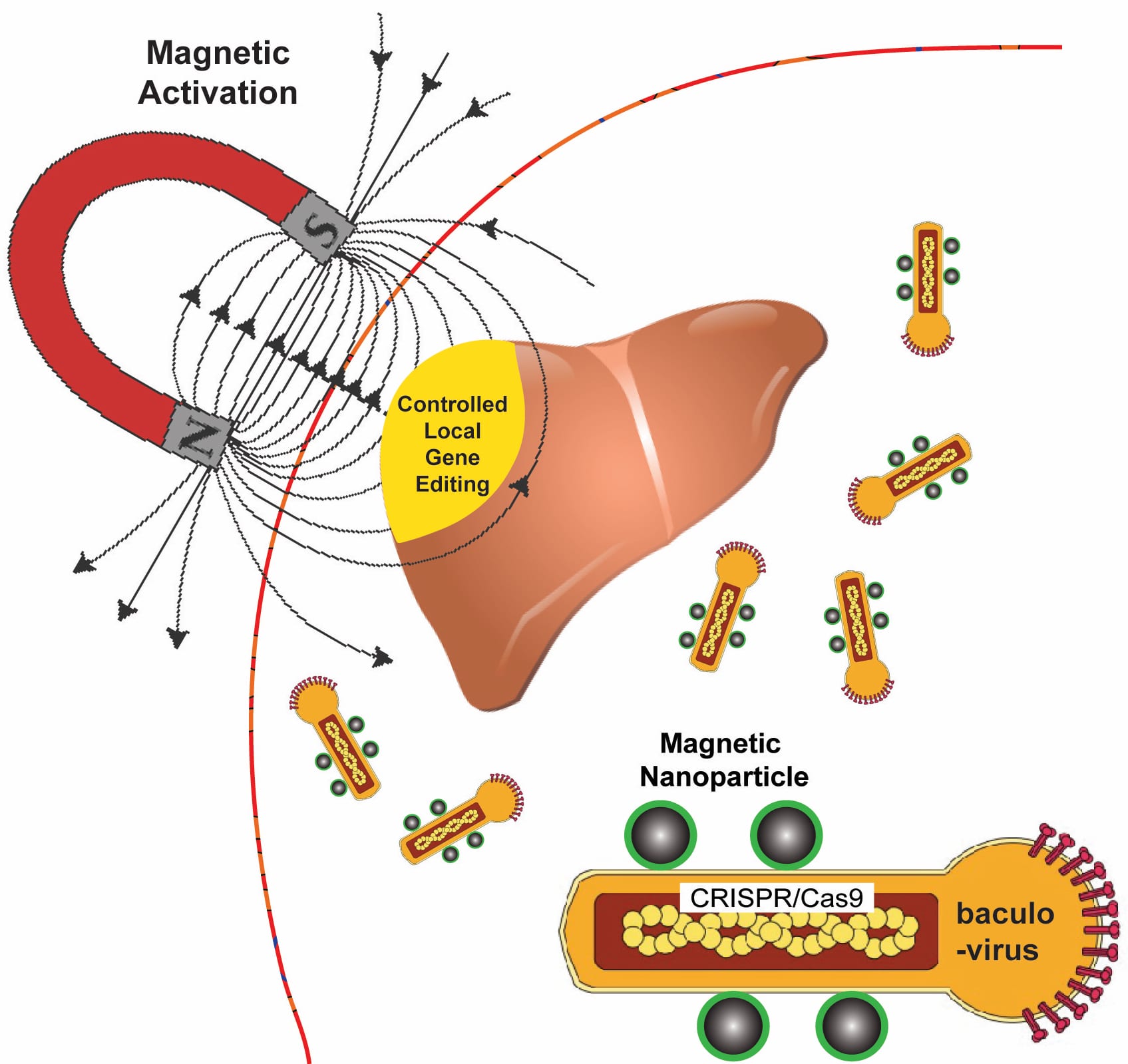
In amidst all the hyperbole about CRISPR (clustered regularly interspaced short palindromic repeats), the gene editing technology, you will sometimes find a mild cautionary note. It seems that CRISPR is not as precise as you might think.
Some months ago there was a story about research into detecting possible unanticipated (off target) effects from using CRISPR, from an April 19, 2019 news item on ScienceDaily,
Since the CRISPR genome editing technology was invented in 2012, it has shown great promise to treat a number of intractable diseases. However, scientists have struggled to identify potential off-target effects in therapeutically relevant cell types, which remains the main barrier to moving therapies to the clinic. Now, a group of scientists at the Gladstone Institutes and the Innovative Genomics Institute (IGI), with collaborators at AstraZeneca, have developed a reliable method to do just that.
…
An April 19, 2019 Gladstone Institutes press release by Julie Langelier, which originated the press release, provides details,
CRISPR edits a person’s genome by cutting the DNA at a specific location. The challenge is to ensure the tool doesn’t also make cuts elsewhere along the DNA—damage referred to as “off-target effects,” which could have unforeseen consequences.
In a study published in the journal Science, the two first authors, Beeke Wienert and Stacia Wyman, found a new way to approach the problem.
“When CRISPR makes a cut, the DNA is broken,” says Wienert, PhD, who began the work in Jacob E. Corn’s IGI laboratory and who is now a postdoctoral scholar in Bruce R. Conklin’s laboratory at Gladstone. “So, in order to survive, the cell recruits many different DNA repair factors to that particular site in the genome to fix the break and join the cut ends back together. We thought that if we could find the locations of these DNA repair factors, we could identify the sites that have been cut by CRISPR.”
To test their idea, the researchers studied a panel of different DNA repair factors. They found that one of them, called MRE11, is one of the first responders to the site of the cut. Using MRE11, the scientists developed a new technique, named DISCOVER-Seq, that can identify the exact sites in the genome where a cut has been made by CRISPR.
“The human genome is extremely large—if you printed the entire DNA sequence, you would end up with a novel as tall as a 16-story building,” explains Conklin, MD, senior investigator at Gladstone and deputy director at IGI. “When we want to cut DNA with CRISPR, it’s like we’re trying to remove one specific word on a particular page in that novel.”
“You can think of the DNA repair factors as different types of bookmarks added to the book,” Conklin adds. “While some may bookmark an entire chapter, MRE11 is a bookmark that drills down to the exact letter than has been changed.”
Different methods currently exist to detect CRISPR off-target effects. However, they come with limitations that range from producing false-positive results to killing the cells they’re examining. In addition, the most common method used to date is currently limited to cultured cells in the laboratory, excluding its use in patient-derived stem cells or animal tissue.
“Because our method relies on the cell’s natural repair process to identify cuts, it has proven to be much less invasive and much more reliable,” says Corn, PhD, who now runs a laboratory at ETH Zurich. “We were able to test our new DISCOVER-Seq method in induced pluripotent stem cells, patient cells, and mice, and our findings indicate that this method could potentially be used in any system, rather than just in the lab.”
The DISCOVER-Seq method, by being applied to new cell types and systems, has also revealed new insights into the mechanisms used by CRISPR to edit the genome, which will lead to a better understanding of the biology of how this tool works.
“The new method greatly simplifies the process of identifying off-target effects while also increasing the accuracy of the results,” says Conklin, who is also a professor of medical genetics and molecular pharmacology at UC San Francisco (UCSF). “This could allow us to better predict how genome editing would work in a clinical setting. As a result, it represents an essential step in improving pre-clinical studies and bringing CRISPR-based therapies closer to the patients in need.”
###
About the Study
The paper “Unbiased detection of CRISPR off-targets in vivo 1 using DISCOVER-Seq” was published by the journal Science on April 19, 2019. Gladstone’s Hannah L. Watry and Luke M. Judge (who is also at UCSF) contributed to this study. Other authors also include Christopher D. Richardson, Jonathan T. Vu, and Katelynn R. Kazane from IGI, Charles D. Yeh from ETH Zurich, as well as Pinar Akcakaya, Michelle J. Porritt, and Michaela Morlock from AstraZeneca.
The work was supported by Gladstone, the National Institutes of Health (grants EY028249 and HL13535801), the Li Ka Shing Foundation, the Heritage Medical Research Institute, the Fanconi Anemia Research Foundation, a Sir Keith Murdoch Fellowship from the American Australian Association, and an Early Career Fellowship from the National Health and Medical Research Council.
About the Gladstone InstituteTo ensure our work does the greatest good, the Gladstone Institutes focuses on conditions with profound medical, economic, and social impact—unsolved diseases. Gladstone is an independent, nonprofit life science research organization that uses visionary science and technology to overcome disease. It has an academic affiliation with the University of California, San Francisco.
Before getting to the link and citation that I usually offer you might find this July 17, 2018 posting, The CRISPR ((clustered regularly interspaced short palindromic repeats)-CAS9 gene-editing technique may cause new genetic damage kerfuffle of interest. I wonder if this latest news affected the CRISPR market as the did the news in 2018.
In addition to the link in the press release, I am including a link and a citation for the study,
Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq by Beeke Wienert, Stacia K. Wyman, Christopher D. Richardson, Charles D. Yeh, Pinar Akcakaya, Michelle J. Porritt, Michaela Morlock, Jonathan T. Vu, Katelynn R. Kazane, Hannah L. Watry, Luke M. Judge, Bruce R. Conklin, Marcello Maresca, Jacob E. Corn. Science 19 Apr 2019: Vol. 364, Issue 6437, pp. 286-289 DOI: 10.1126/science.aav9023
This paper is behind a paywall.
Money
Over the last 10 or more years, I have, on occasion made a point, of finding out about the funding for various non-profit agencies and projects. I find that sort of thing interesting and have hoped that my readers might feel the same way.
It seems that my readers and I might not be the only ones to care about the source of funding. Joi Ito who held appointments with Harvard University and the Massachusetts Institute of Technology (MIT) resigned from his various appointments on Sept. 7, 2019 after news of major donations from Jeffrey Epstein (a disgraced financier and sex offender) to MIT were revealed. From the Joi Ito’s entry on Wikipedia (Note: Links have been removed),
Joichi “Joi” Ito (伊藤 穰一 Itō Jōichi, born June 19, 1966) is a Japanese activist, entrepreneur and venture capitalist. He is the former director of the MIT Media Lab, and a former professor of the practice of media arts and sciences at MIT. He is a former visiting professor of practice at the Harvard Law School.[1][2]
Ito has received recognition for his role as an entrepreneur focused on Internet and technology companies and has founded, among other companies, PSINet Japan, Digital Garage and Infoseek Japan. Ito is a strategic advisor to Sony Corporation[3] and general partner of Neoteny Labs.[4] Ito writes a monthly column in the Ideas section of Wired.[5]
Ito resigned from his roles at MIT, Harvard, the John D. and Catherine T. MacArthur Foundation, the Knight Foundation, PureTech Health and The New York Times Company on September 7, 2019, following allegations of financial ties to sex offender and financier Jeffrey Epstein.[2][6][7]
…
Many, many institutions have accepted funds from sketchy characters and orgnaizations. It’s not new to academia, the sciences, or the arts. For a contemporary view of how some of this works, take a look at Anand Giridharadas’s 2018 book, Winners Take All. From the webepage for the book,
WINNERS TAKE ALL
The Elite Charade of Changing the World
An insider’s groundbreaking investigation of how the global elite’s efforts to “change the world” preserve the status quo and obscure their role in causing the problems they later seek to solve.
Former New York Times columnist Anand Giridharadas takes us into the inner sanctums of a new gilded age, where the rich and powerful fight for equality and justice any way they can–except ways that threaten the social order and their position atop it. We see how they rebrand themselves as saviors of the poor; how they lavishly reward “thought leaders” who redefine “change” in winner-friendly ways; and how they constantly seek to do more good, but never less harm. We hear the limousine confessions of a celebrated foundation boss; witness an American president hem and haw about his plutocratic benefactors; and attend a cruise-ship conference where entrepreneurs celebrate their own self-interested magnanimity.…
I don’t recall any mention of Epstein in Giridharadas’s book but he did have this to say on Twitter about Epstein,
Anand GiridharadasVerified account @AnandWrites
Everything that made Epstein’s life possible remains in place after his arrest: the Caribbean tax havens, the hidden real-estate deals, the buying of politicians, the nonprofits that sell reputational glow, the editors who cover for people of their class.7:34 PM – 8 Jul 2019
it can’t be easy to withstand the temptation to take the money and hope that the misdoings have been exaggerated or that they have stopped. I imagine Ito and others are under constant pressure to get funds.
AstraZeneca
One of the partners in this research about CRISPR, AstraZeneca, is a pharmaceutical company. In fact, it’s one of the largest in the world (from the AstraZeneca Wikipedia entry; Note: Links have been removed),
AstraZeneca plc[4] is a British-Swedish multinational pharmaceutical and biopharmaceutical company. In 2013, it moved its headquarters to Cambridge, UK, and concentrated its R&D in three sites: Cambridge; Gaithersburg, Maryland, USA (location of MedImmune) for work on biopharmaceuticals; and Mölndal (near Gothenburg) in Sweden, for research on traditional chemical drugs.[5] AstraZeneca has a portfolio of products for major disease areas including cancer, cardiovascular, gastrointestinal, infection, neuroscience, respiratory and inflammation.[6]
The company was founded in 1999 through the merger of the Swedish Astra AB and the British Zeneca Group[7][8] (itself formed by the demerger of the pharmaceutical operations of Imperial Chemical Industries in 1993). Since the merger it has been among the world’s largest pharmaceutical companies and has made numerous corporate acquisitions, including Cambridge Antibody Technology (in 2006), MedImmune (in 2007), Spirogen (in 2013) and Definiens (by MedImmune in 2014).
…
ControversiesSeroquel
In April 2010 AstraZeneca settled a qui tam lawsuit brought by Stefan P. Kruszewski for $520 million to settle allegations that the company defrauded Medicare, Medicaid, and other government-funded health care programs in connection with its marketing and promotional practices for the blockbuster atypical antipsychotic, Seroquel.[76]
In March 2011, AstraZeneca settled a lawsuit in the United States totalling $68.5 million to be divided up to 38 states.[77]
Nexium
The company’s most commercially successful medication is esomeprazole (Nexium). The primary uses are treatment of gastroesophageal reflux disease, treatment and maintenance of erosive esophagitis, treatment of duodenal ulcers caused by Helicobacter pylori, prevention of gastric ulcers in those on chronic NSAID therapy, and treatment of gastrointestinal ulcers associated with Crohn’s disease. When it is manufactured the result is a mixture of two mirror-imaged molecules, R and S. Two years before the omeprazole patent expired, AstraZeneca patented S-omeprazole in pure form, pointing out that since some people metabolise R-omeprazole slowly, pure S-omeprazole treatment would give higher dose efficiency and less variation between individuals.[78] In March 2001, the company began to market Nexium, as it would a brand new drug.[79]
…
In 2007, Marcia Angell, former editor-in-chief of the New England Journal of Medicine and a lecturer in social medicine at the Harvard Medical School, said in Stern, a German-language weekly newsmagazine, that AstraZeneca’s scientists had misrepresented their research on the drug’s efficiency, saying “Instead of using presumably comparable doses [of each drug], the company’s scientists used Nexium in higher dosages. They compared 20 and 40 mg Nexium with 20 mg Prilosec. With the cards having been marked in that way, Nexium looked like an improvement – which however was only small and shown in only two of the three studies.”[83]
Bildman fraud, and faithless servant clawback
…
Study
In 2004, University of Minnesota research participant Dan Markingson committed suicide while enrolled in an industry-sponsored pharmaceutical trial comparing three FDA-approved atypical antipsychotics: Seroquel (quetiapine), Zyprexa (olanzapine), and Risperdal (risperidone). University of Minnesota Professor of Bioethics Carl Elliott noted that Markingson was enrolled in the study against the wishes of his mother, Mary Weiss, and that he was forced to choose between enrolling in the study or being involuntarily committed to a state mental institution.[89] Further investigation revealed financial ties to AstraZeneca by Markingson’s psychiatrist, Stephen C. Olson, oversights and biases in AstraZeneca’s trial design, and the inadequacy of university Institutional Review Board (IRB) protections for research subjects.[90][unreliable source?] A 2005 FDA investigation cleared the university. Nonetheless, controversy around the case has continued. A Mother Jones article[89] resulted in a group of university faculty members sending a public letter to the university Board of Regents urging an external investigation into Markingson’s death.[91]
…
Is it ok to take money and/or other goods and services from them?
Innovative Genomics Institute (IGI)
Also mentioned as a partner in the research, is the Innovative Genomics Institute (IGI). Here’s more from the company’s Overview webpage (Note: Links have been removed),,
The IGI began in 2014 through the Li Ka Shing Center for Genetic Engineering, which was created thanks to a generous donation from the Li Ka Shing Foundation. [emphasis mine] The Innovative Genomics Initiative formed as a partnership between the University of California, Berkeley and the University of California, San Francisco. Combining the fundamental research expertise and the biomedical talent at UCB and UCSF, the Innovative Genomics Initiative focused on unraveling the mechanisms underlying CRISPR-based genome editing and applying this technology to improve human health. Early achievements include improving the efficiency of gene replacement and foundational work toward a treatment for sickle cell disease.
In late 2015, generous philanthropic donations enabled a bolder vision and broader mission for the IGI. With this expansion came a significant enhancement of the organization, and in January 2017, the IGI officially re-launched as the Innovative Genomics Institute.
…
As it turns out, there is a Li Ka-shing and he has a bit of a history with Vancouver (Canada). First, here’s more about him from the Li Ka-shing Wikipedia entry,(Note: Links have been removed),
Sir Li Ka-shing GBM KBE JP[4] (born 13 June 1928)[5][6] is a Hong Kong business magnate, investor, and philanthropist. As of June 2019, Li is the 30th richest person in the world, with an estimated net wealth of US$29.4 billion.[3] He is the senior advisor for CK Hutchison Holdings,[7] after he retired from the Chairman of the Board in May 2018;[8] through it, he is the world’s leading port investor, developer, and operator of the largest health and beauty retailer in Asia and Europe.[9]
…
Besides business through his flagship companies Cheung Kong Property Holdings and CK Hutchison Holdings Limited, Li Ka-shing has also personally invested extensively in real estate in Singapore and Canada. He was the single largest shareholder of Canadian Imperial Bank of Commerce (CIBC), the fifth largest bank in Canada, until the sale of his share in 2005 (with all proceedings donated, see below). He is also the majority shareholder of a major energy company, Husky Energy, based in Alberta, Canada.[48]
In January 2005, Li announced plans to sell his $1.2 billion CAD stake in the Canadian Imperial Bank of Commerce, with all proceeds going to private charitable foundations established by Li, including the Li Ka Shing Foundation in Hong Kong and the Li Ka Shing (Canada) Foundation based in Toronto, Ontario.[49]
…
His son Victor Li was kidnapped in 1996 on his way home after work by gangster “Big Spender” Cheung Tze-keung. Li Ka-shing paid a ransom of HK$1 billion, directly to Cheung who had come to his house.[53] A report was never filed with Hong Kong police. Instead the case was pursued by Mainland authorities, leading to Cheung’s execution in 1998, an outcome not possible under Hong Kong law. Rumours circulated of a deal between Li and the Mainland.[53] In interviews, when this rumor was brought up, Li brushed it off and dismissed it completely.
…
Li Ka-shing was well known here in Vancouver due to his purchase of a significant chunk of land in the city. This January 9, 2015 article by Glen Korstrum for Business in Vancouver notes some rather interesting news and contextualizes with Li’s Vancouver history,
Hong Kong billionaire Li Ka-shing is restructuring his empire and shifting his base to the Cayman Islands and away from the Chinese special administrative region.
His January 9 [2015] announcement came the same day that Forbes ranked him as Hong Kong’s richest man for the 17th consecutive year, with a total wealth of US$33.5 billion.
Li is best known in Vancouver for buying an 82.5-hectare parcel of land around False Creek for $328 million in 1988 along with partners, who included fellow Hong Kong tycoons, Lee Shau Kee and Cheng Yu Tung.
The group formed Concord Pacific, which redeveloped the site that had been home to Vancouver’s 1986 world’s fair, Expo ’86.
Li cashed out of Concord Pacific in the late 1990s and, in 2007, invested in Deltaport through his Hutchison Port Holdings.Li’s biggest Canadian holding is his controlling stake in Husky Energy. …
Intriguing, yes? It also makes the prospect of deciding whose money you’re going to accept a bit more complicated than it might seem.
Gladstone Institutes
In what seems to be a decided contrast to the previous two partners, here’s more from the Gladstone Institutes, About Us, History webpage,
Born in London in 1910, J. David Gladstone was orphaned as a boy and came to North America at age 10. He began a career in real estate in Southern California at age 28, eventually making his fortune as the first developer to create the region’s enclosed shopping malls (such as the Northridge Fashion Center mall). His accidental death in 1971 left an estate valued at about $8 million to support medical students interested in research.
It soon became clear to the three trustees administering Mr. Gladstone’s trust that his legacy could support a far more substantial philanthropic enterprise. In 1979, they launched The J. David Gladstone Institutes under the leadership of Robert W. Mahley, MD, PhD, a leading cardiovascular scientist who at the time was working at the National Institutes of Health.
…
In 2010, after three decades of leading Gladstone, Dr. Mahley stepped down in order to return to more active research. That same year, R. Sanders “Sandy” Williams, MD, left Duke University, where he had been Dean of the School of Medicine—as well as Senior Vice Chancellor and Senior Advisor for International Strategy—to become Gladstone’s new president. The following year, the S.D. Bechtel, Jr. Foundation [emphasis mine] helped launch the Center for Comprehensive Alzheimer’s Disease Research with a generous $6M lead gift, while the Roddenberry Foundation [emphasis mine] gave $5 million to launch the Roddenberry Center for Stem Cell Biology and Medicine. Also in 2011, the independent and philanthropic Gladstone Foundation formed with the mission of expanding the financial resources available to drive’s Gladstone’s mission.
…
The S. D. Bechtel jr. mentioned is associated with Bechtel, an international engineering firm. I did not find any scandals or controversies in the Bechtel Wikipedia entry. That seemed improbable so I did a little digging and found a January 30, 2015 (?) article by Matthew Brunwasser for foreignpolicy.com (Note: A link has been removed),
Steamrolled; A special investigation into the diplomacy of doing business abroad.
One of Europe’s poorest countries wanted a road, so U.S. mega-contractor Bechtel sold it a $1.3 billion highway, with the backing of a powerful American ambassador. Funny thing is, the highway is barely being used—and the ambassador is now working for Bechtel.
…
Bechtel, the largest contractor by revenue in the United States and the third-largest internationally, according to an annual list compiled by the Engineering News-Record, has in recent years constructed expensive highways in Kosovo, Croatia, Romania, and Albania. A six-month investigation by the Investigative Reporting Program at the University of California at Berkeley Graduate School of Journalism has found that these highways were boondoggles for the countries in which they were constructed, and that members of governments and international institutions often saw problems coming before Bechtel (along with its Turkish joint venture partner, Enka) even began work on the roads.
…
My other source is a May 8, 1988 article by Walter Russell Mead for the Los Angeles Time,s
From San Francisco to Saudi Arabia, the Bechtel Group Inc. has left its mark around the world. Yet the privately owned Bechtel Group is one of the country’s most mysterious operations–or was, until the publication of Laton McCartney’s critical and controversial “Friends in High Places.”
Those who believe that “Dynasty” and “Falcon Crest” describe life at the top of America’s corporate pyramids will find a picture here that makes the most far-fetched TV plots look dull. One Bechtel executive was torn to pieces by an angry mob; another, kidnaped, survived two days in the trunk of a Mercedes that had been driven over the edge of a cliff but caught on an obstacle half way down. Wheeling and dealing from Beirut to the Bohemian Grove, Bechtel executives fought off Arab and Jewish nationalists, angry senators, bitter business rivals, and furious consumer groups to build the world’s largest construction and engineering firm.
…
Poor Bechtel sometimes seems damned if it does and damned if it doesn’t. No major corporation could undertake foreign operations on Bechtel’s scale without some cooperation from the U.S. government–and few companies could refuse a government request that, in return, they provide cover for intelligence agents. Given the enormous scope of Bechtel’s operations in global trouble spots–a $20-billion industrial development in Saudi Arabia, for example–it could only proceed with assurances that its relations with both Saudi and American governments were good. Where, exactly, is the line between right and wrong? [emphasis mine]
…
… The white elephants Bechtel scattered across the American landscape–particularly the nuclear power plants that threaten to bankrupt some of the country’s largest utility systems–are monuments to wasted talent and misdirected resources.
…
Finally, I get to the Roddenberry Foundation, which was founded by Gene Roddenberry’s (Star Trek) son. Here’s more from the About Us, Origin webpage,
Gene Roddenberry, creator of the Star Trek series, brought to his audiences meaningful and thought-provoking science fiction to “think, question, and challenge the status quo” with the intention of creating “a brighter future”. His work has touched countless lives and continues to entertain and inspire audiences worldwide. In 2010, Gene’s son Rod established the Roddenberry Foundation to build on his father’s legacy and philosophy of inclusion, diversity, and respect for life to drive social change and meaningfully improve the lives of people around the world.
…
While there are many criticisms of Mr. Roddenberry, there doesn’t seem to be anything that would be considered a serious scandal on the order of a Jeffrey Epstein or the whisper of scandal on the order of Sir Li Ka-shing or Bechtel.
Final comments
It’s a good thing when research is funded and being able to detect off-target effects from CRISPR is very good, assuming the research holds up to closer scrutiny.
As for vetting your donors, that’s tricky. Of course, Epstein was already a convicted sex offender when Ito accepted his funding for MIT but I cannot emphasize enough the amount of pressure these folks are under. Academia is always hungry for money. Hopefully this incident will introduce checks and balances in the donor process.