Markus Buehler and his musical spider webs are making news again.
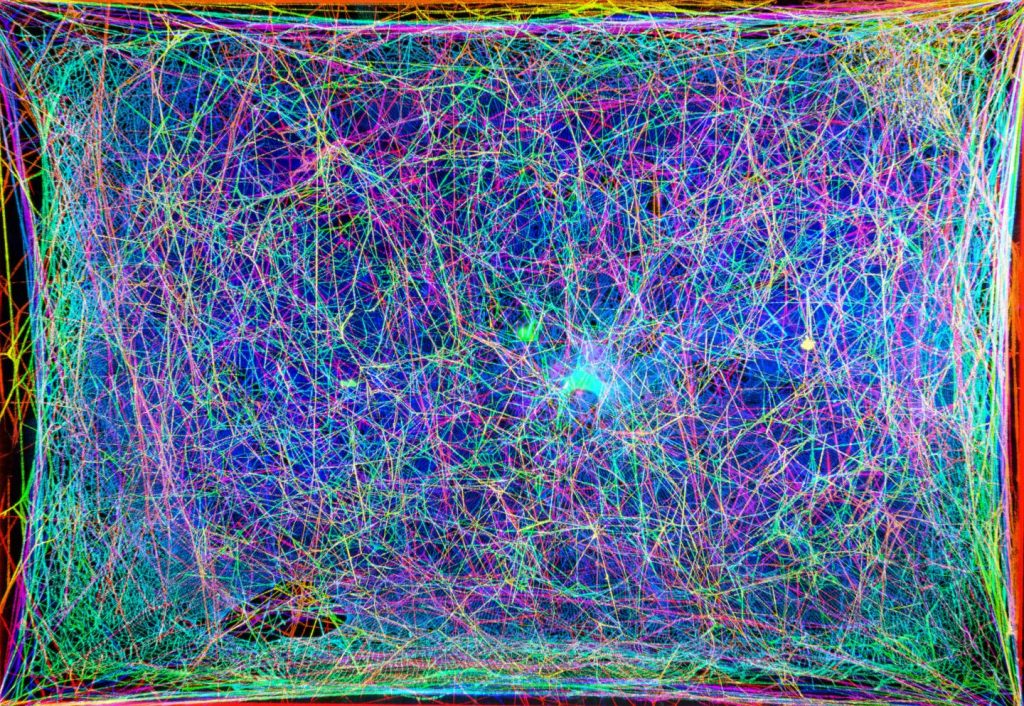
The image (so pretty) you see in the above comes from a Markus Buehler presentation that was made at the American Chemical Society (ACS) meeting. ACS Spring 2021 being held online April 5-30, 2021. The image was also shown during a press conference which the ACS has made available for public viewing. More about that later in this posting.
The ACS issued an April 12, 2021 news release (also on EurekAlert), which provides details about Buehler’s latest work on spider webs and music,
Spiders are master builders, expertly weaving strands of silk into intricate 3D webs that serve as the spider’s home and hunting ground. If humans could enter the spider’s world, they could learn about web construction, arachnid behavior and more. Today, scientists report that they have translated the structure of a web into music, which could have applications ranging from better 3D printers to cross-species communication and otherworldly musical compositions.
The researchers will present their results today at the spring meeting of the American Chemical Society (ACS). ACS Spring 2021 is being held online April 5-30 [2021]. Live sessions will be hosted April 5-16, and on-demand and networking content will continue through April 30 [2021]. The meeting features nearly 9,000 presentations on a wide range of science topics.
“The spider lives in an environment of vibrating strings,” says Markus Buehler, Ph.D., the project’s principal investigator, who is presenting the work. “They don’t see very well, so they sense their world through vibrations, which have different frequencies.” Such vibrations occur, for example, when the spider stretches a silk strand during construction, or when the wind or a trapped fly moves the web.
Buehler, who has long been interested in music, wondered if he could extract rhythms and melodies of non-human origin from natural materials, such as spider webs. “Webs could be a new source for musical inspiration that is very different from the usual human experience,” he says. In addition, by experiencing a web through hearing as well as vision, Buehler and colleagues at the Massachusetts Institute of Technology (MIT), together with collaborator Tomás Saraceno at Studio Tomás Saraceno, hoped to gain new insights into the 3D architecture and construction of webs.
With these goals in mind, the researchers scanned a natural spider web with a laser to capture 2D cross-sections and then used computer algorithms to reconstruct the web’s 3D network. The team assigned different frequencies of sound to strands of the web, creating “notes” that they combined in patterns based on the web’s 3D structure to generate melodies. The researchers then created a harp-like instrument and played the spider web music in several live performances around the world.
The team also made a virtual reality setup that allowed people to visually and audibly “enter” the web. “The virtual reality environment is really intriguing because your ears are going to pick up structural features that you might see but not immediately recognize,” Buehler says. “By hearing it and seeing it at the same time, you can really start to understand the environment the spider lives in.”
To gain insights into how spiders build webs, the researchers scanned a web during the construction process, transforming each stage into music with different sounds. “The sounds our harp-like instrument makes change during the process, reflecting the way the spider builds the web,” Buehler says. “So, we can explore the temporal sequence of how the web is being constructed in audible form.” This step-by-step knowledge of how a spider builds a web could help in devising “spider-mimicking” 3D printers that build complex microelectronics. “The spider’s way of ‘printing’ the web is remarkable because no support material is used, as is often needed in current 3D printing methods,” he says.
In other experiments, the researchers explored how the sound of a web changes as it’s exposed to different mechanical forces, such as stretching. “In the virtual reality environment, we can begin to pull the web apart, and when we do that, the tension of the strings and the sound they produce change. At some point, the strands break, and they make a snapping sound,” Buehler says.
The team is also interested in learning how to communicate with spiders in their own language. They recorded web vibrations produced when spiders performed different activities, such as building a web, communicating with other spiders or sending courtship signals. Although the frequencies sounded similar to the human ear, a machine learning algorithm correctly classified the sounds into the different activities. “Now we’re trying to generate synthetic signals to basically speak the language of the spider,” Buehler says. “If we expose them to certain patterns of rhythms or vibrations, can we affect what they do, and can we begin to communicate with them? Those are really exciting ideas.”
You can go here for the April 12, 2021 ‘Making music from spider webs’ ACS press conference’ it runs about 30 mins. and you will hear some ‘spider music’ played.
Getting back to the image and spider webs in general, we are most familiar with orb webs (in the part of Canada where I from if nowhere else), which look like spirals and are 2D. There are several other types of webs some of which are 3D, like tangle webs, also known as cobwebs, funnel webs and more. See this March 18, 2020 article “9 Types of Spider Webs: Identification + Pictures & Spiders” by Zach David on Beyond the Treat for more about spiders and their webs. If you have the time, I recommend reading it.
I’ve been following Buehler’s spider web/music work for close to ten years now; the latest previous posting is an October 23, 2019 posting where you’ll find a link to an application that makes music from proteins (spider webs are made up of proteins; scroll down about 30% of the way; it’s in the 2nd to last line of the quoted text about the embedded video).
Here is a video (2 mins. 17 secs.) of a spider web music performance that Buehler placed on YouTube,
Feb 3, 2021
Markus J. Buehler
Spider’s Canvas/Arachonodrone show excerpt at Palais de Tokyo, Paris, on November 2018. Video by MIT CAST. More videos can be found on www.arachnodrone.com. The performance was commissioned by Studio Tomás Saraceno (STS), in the context of Saraceno’s carte blanche exhibition, ON AIR. Spider’s Canvas/Arachnodrone was performed by Isabelle Su and Ian Hattwick on the spider web instrument, Evan Ziporyn on the EWI (Electronic Wind Instrument), and Christine Southworth on the guitar and EBow (Electronic Bow)
You can find more about the spider web music and Buehler’s collaborators on http://www.arachnodrone.com/,
Spider’s Canvas / Arachnodrone is inspired by the multifaceted work of artist Tomas Saraceno, specifically his work using multiple species of spiders to make sculptural webs. Different species make very different types of webs, ranging not just in size but in design and functionality. Tomas’ own web sculptures are in essence collaborations with the spiders themselves, placing them sequentially over time in the same space, so that the complex, 3-dimensional sculptural web that results is in fact built by several spiders, working together.
Meanwhile, back among the humans at MIT, Isabelle Su, a Course 1 doctoral student in civil engineering, has been focusing on analyzing the structure of single-species spider webs, specifically the ‘tent webs’ of the cyrtophora citricola, a tropical spider of particular interest to her, Tomas, and Professor Markus Buehler. Tomas gave the department a cyrtophora spider, the department gave the spider a space (a small terrarium without glass), and she in turn built a beautiful and complex web. Isabelle then scanned it in 3D and made a virtual model. At the suggestion of Evan Ziporyn and Eran Egozy, she then ported the model into Unity, a VR/game making program, where a ‘player’ can move through it in numerous ways. Evan & Christine Southworth then worked with her on ‘sonifying’ the web and turning it into an interactive virtual instrument, effectively turning the web into a 1700-string resonating instrument, based on the proportional length of each individual piece of silk and their proximity to one another. As we move through the web (currently just with a computer trackpad, but eventually in a VR environment), we create a ‘sonic biome’: complex ‘just intonation’ chords that come in and out of earshot according to which of her strings we are closest to. That part was all done in MAX/MSP, a very flexible high level audio programming environment, which was connected with the virtual environment in Unity. Our new colleague Ian Hattwick joined the team focusing on sound design and spatialization, building an interface that allowed him the sonically ‘sculpt’ the sculpture in real time, changing amplitude, resonance, and other factors. During this performance at Palais de Tokyo, Isabelle toured the web – that’s what the viewer sees – while Ian adjusted sounds, so in essence they were together “playing the web.” Isabelle provides a space (the virtual web) and a specific location within it (by driving through), which is what the viewer sees, from multiple angles, on the 3 scrims. The location has certain acoustic potentialities, and Ian occupies them sonically, just as a real human performer does in a real acoustic space. A rough analogy might be something like wandering through a gothic cathedral or a resonant cave, using your voice or an instrument at different volumes and on different pitches to find sonorous resonances, echoes, etc. Meanwhile, Evan and Christine are improvising with the web instrument, building on Ian’s sound, with Evan on EWI (Electronic Wind Instrument) and Christine on electric guitar with EBow.
For the visuals, Southworth wanted to create the illusion that the performers were actually inside the web. We built a structure covered in sharkstooth scrim, with 3 projectors projecting in and through from 3 sides. Southworth created images using her photographs of local Lexington, MA spider webs mixed with slides of the scan of the web at MIT, and then mixed those images with the projection of the game, creating an interactive replica of Saraceno’s multi-species webs.
If you listen to the press conference, you will hear Buehler talk about practical applications for this work in materials science.